Introduction
We are especially interested – and have been for a quarter of a century (Kell, D.B. (1979) BBA 549, 55-99) – in the holistic analysis of complex systems. This is now often referred to as systems biology. Here we are setting out some views of Systems Biology.
"So the first requirement will be for a theoretical framework in which to embed all the detailed knowledge we have accumulated, to allow us to compute outcomes of the complex interactions and to start to understand the dynamics of the system. The second will be to make parallel measurements of the behaviour of many components during the execution by the cell of an integrated action in order to test whether the theory is right. Is there some other approach? If I knew it I would be doing it, and not writing about the problem."
Sydney Brenner, in Loose Ends, Current Biology, 1997, p. 73
"…an approach that is inefficient in analyzing a simple system is unlikely to be more useful if the system is more complex." (Lazebnik, 2002)
In an amusing and well-done article (can a biologist fix a radio?), Lazebnik (2002) points out that despite the floods of 'data' (some 10,000 papers per year on apoptosis in the last few years, and more than 23,000 on p53 alone) we are little nearer understanding cell function. (Of curse we still do not know the 'function(s)' of half the genes in even well-worked organisms like S. cerevisiae and E. coli.) He contrasts the approach of the molecular biologist with that of an engineer. A molecular biology postdoc would remove a component (i.e. 'knock out a gene') from the radio and if the radio failed to work (make music) the component might be named Serendipitously Recovered Component (Src) and a glowing career assured. Soon another postdoc will come along and find a Most Important Component (Mic) or really important component (Ric) which have similar 'importance' to the functioning of the radio, and so on.
Of course what matters for the radio – as well as for the cell or organism – is not only what is there but, perhaps more importantly, how they are connected up, and the engineer would want to see the circuit diagram, with which one might expect to understand how the radio works. An engineer would not believe that putting the radio in a blender, centrifuging the pieces to separate them on the basis of their relative density and then further separating them on a 2D gel according to mass and charge would tell him or her how this complex system worked, even if a molecular biologist might.
Although Systems Biology does not inherently imply the sciences of 'complexity' (e.g. ( Kell & Welch, 1991; Axelrod & Cohen, 1999; Schirmer, 1995; Coveney & Highfield, 1995; Oltvai & Barabasi, 2002; Csete & Doyle, 2002) for a sampling) or 'chaos' ((Gleick, 1988) for a popular introduction), most systems in which we are interested are in fact sufficiently 'complex' (with nonlinearities, emergent properties, loosely coupled modules, etc. that are the hallmarks of 'complexity') that we are wise to bear in mind that 'systems biology' and 'complex systems' are in fact practically synonymous. Tools found useful in analysing the latter will prove of value to the study of the former.
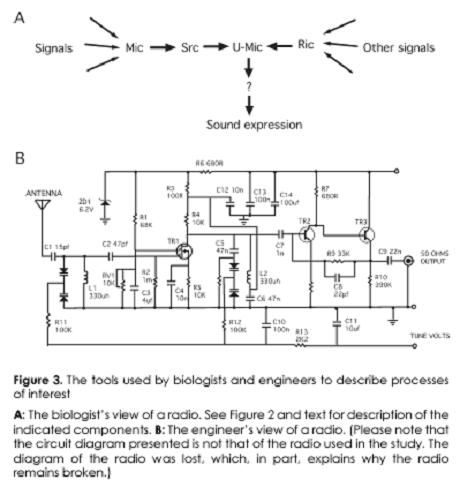
From (Lazebnik, 2002). Note the presence of numerical values in B and their absence in A.
So to understand a system, we first have to have a so-called "structural model" (this terminology is nothing to do with 'structural biology' or 'structural genomics' (Brenner, 2001) in the sense of the 3D coordinates of atoms in a molecule), by which is meant a wiring diagram which shows all the components and, qualitatively, how they talk to each other. Then we have to understand in quantitative terms how these links behave, so we can base the model on equations with relationships that can reflect reality, and then parametrise them. This is typically done by perturbing the system in a specified way, and looking at the time evolution of the system, and ultimately solving the inverse problem (given the measured variables, estimate the parameters). However, such problems are often under-determined or ill-conditioned, and competing models may be hard to distinguish. But only then can we begin to understand the properties of the system, and 'systems biology' is seen, significantly, to be the science of analysing and modelling genetic, macromolecular and metabolic networks.
We can discriminate a variety of strategies for understanding complex systems (which may be large or small but will normally be nonlinear, and often highly so). One is the directional relationship between the whole and parts, another between the world of ideas/hypotheses and the world of data/observations. Often these are contrasted, but I consider that they are best viewed as cycles. Systems Biology – or integrative biology – requires this iteration between data-driven science, hypothesis generation and experimentation (Kell & Oliver, 2003).
There are many different definitions of Systems Biology; like an elephant it is easy to recognise and hard to define. Henry (2003) gives an overview. A hopefully uncontroversial definition would be "Integrative approaches in which scientists study and model pathways and networks, with an interplay between experiment and theory". Kitano's version ( Kitano, 2002a; Kitano, 2002b) reads "Systems biology has two distinct branches: Knowledge discovery and data mining, which extract the hidden pattern from huge quantities of experimental data, forming hypothesis as result and simulation-based analysis, providing predictions to be tested by in vitro and in vivo studies." Westerhoff, cited in (Henry, 2003), stresses the emergent properties of networks, that which is 'in-between' the parts and the whole. Leroy Hood's titular definition (Hood, 2003) involves 'integrating technology, biology and computation'.
The hallmarks of systems biology certainly include the omics methods with which we shall populate our models, and thus tools and technologies connected with large-scale measurements at one or (preferably) more levels of biological organisation are crucial to systems biology. How much this is about technology development as opposed to usage and validation of methods (including statistically), and how this fits in with other parts of the strategy needs looking at. Most current expression profiling 'omics' methods are done on cell extracts, which means that we lose the information about how things are actually organised in vivo; progress on in vivo measurements is highly desirable.
A clearly discernable and desirable trend in our Integrative Biology is to seek to study systems at different levels of organisation (genomic, transcriptomic, proteomic, metabolomic, phenotypic) and develop methods for inferring causal relationships between the expression profiles or properties at the different levels. This needs promoting.
A crucial part of systems biology is quantitative modelling, for instance using ODE solving systems such as Gepasi (Mendes, 1997; Mendes & Kell, 2001) and MEG (Mendes & Kell, 2001). The 'in silico' cell is thus a very desirable goal, since a key recognition of Systems Biology is that modelling is a substantial part of understanding and 'identifying' complex systems. The ability to exchange models (including their metadata) requires suitable mark-up languages such as the Systems Biology Markup Language (SBML).
Although it is commonplace in the physical sciences and engineering (the Boeing 777 was developed entirely in silico before it was tested in wind tunnels etc 'for real'), it is now recognised as desirable to produce a mathematical model of the system in which one is interested (Mendes & Kell, 1998; Westerhoff & Kell, 1996; Schilling et al., 1999; Tomita et al., 1999; Giersch, 2000; Ouzounis & Karp, 2000; Teusink et al., 2000; Gombert & Nielsen, 2000; Hofmeyr & Westerhoff, 2001; Noble, 2002a; Noble, 2002b; Hoffmann et al., 2002; Mendes, 2002), since such models have at least the following beneficial properties: (i) they serve to provide a mathematical underpinning of our knowledge and its internal consistency, (ii) they allow us to do 'what if?' experiments in silico to determine, for instance, whether a specific intervention will have the desired (or indeed unexpected) effects 'downstream', as in metabolic engineering (Kell & Westerhoff, 1986; Bailey, 1991) and pharmaceutical target validation (Hughes et al., 2000), (iii) they allow one to perform sensitivity analysis to determine which parameters of the system are responsible for the bulk of the control of properties such as metabolic fluxes.
I hesitate to use the word 'bioinformatics', but the non-modelling part – as well, indeed, as the modelling part – will need good integrated data resources that bring together genomic, expression profiling and functional/phenotypic data with the methods of clustering and machine learning. Only then can we move to an integrative biology. Visualisation is another area where we need progress. It needs to be integrated with the data reduction and exploratory data analysis.
Strongly related to Systems Biology is 'network science', exemplified in concepts such as 'scale-free' (Barabasi, 2002) and 'small-world' (Watts & Strogatz, 1998; Wagner & Fell, 2001) networks, network motifs (Shen-Orr et al., 2002; Milo et al., 2002), modularity (Ravasz et al., 2002), robustness (von Dassow et al., 2000) and so on. 'Network properties' that are likely to be of general interest include switching, signal amplification, distribution of control and robustness. There is likely to be an interplay between modelling and bioinformatics here. See the BBSRC 10-year vision for further examples.
Update Jan 2007: More recent summaries can be found in Kell 2006 a,b,c all accessible from my Publications page. See also links from MCISB.
| Publications |
|---|
|
Lazebnik, Y. (2002). Can a biologist fix a radio? – or, what I learned while studying apoptosis. Cancer Cell 2, 179-182. Kell, D. B. & Welch, G. R. (1991). No turning back, Reductonism and Biological Complexity. Times Higher Educational Supplement 9th August, 15. Axelrod, R. & Cohen, M. D. (1999). Harnessing complexity: organisational implications of a scientific frontier. The Free Press, New York. Schirmer, A. (1995). A guide to complexity theory in operations research. Manuskripte aus den Instituten fur Betriebswirtschaftslehre der Universitat Kiel 381. Coveney, P. V. & Highfield, R. R. (1995). Frontiers of complexity. Faber & Faber, London. Oltvai, Z. N. & Barabasi, A. L. (2002). Systems biology. Life's complexity pyramid. Science 298, 763-4. Csete, M. E. & Doyle, J. C. (2002). Reverse engineering of biological complexity. Science 295, 1664-1669. Gleick, J. (1988). Chaos: making a new science. Abacus, New York. Brenner, S. E. (2001). A tour of structural genomics. Nat Rev Genet 2, 801-9. Kell, D. B. & Oliver, S. G. (2004). Here is the evidence, now what is the hypothesis? The complementary roles of inductive and hypothesis-driven science in the post-genomic era. Bioessays 26, 99-105. Henry, C. M. (2003). Systems biology. Chem. Eng. News 81, 45-55. Kitano, H. (2002a). Systems biology: a brief overview. Science 295, 1662-4. Kitano, H. (2002b). Computational systems biology. Nature 420, 206-10. Hood, L. (2003). Systems biology: integrating technology, biology, and computation. Mech Ageing Dev 124, 9-16. Mendes, P. & Kell, D. B. (1998). Non-linear optimization of biochemical pathways: applications to metabolic engineering and parameter estimation. Bioinformatics 14, 869-883. Mendes, P. (1997). Biochemistry by numbers: simulation of biochemical pathways with Gepasi 3. Trends Biochem. Sci. 22, 361-363. Mendes, P. & Kell, D. B. (2001). MEG (Model Extender for Gepasi): a program for the modelling of complex, heterogeneous cellular systems. Bioinformatics 17, 288-289. Westerhoff, H. V. & Kell, D. B. (1996). What BioTechnologists knew all along…? J. Theoret. Biol. 182, 411-420. Schilling, C. H., Schuster, S., Palsson, B. O. & Heinrich, R. (1999). Metabolic pathway analysis: Basic concepts and scientific applications in the post-genomic era. Biotechnology Progress 15, 296-303. Tomita, M., Hashimoto, K., Takahashi, K., Shimizu, T. S., Matsuzaki, Y., Miyoshi, F., Saito, K., Tanida, S., Yugi, K., Venter, J. C. & Hutchison, C. A. (1999). E-CELL: software environment for whole-cell simulation. Bioinformatics 15, 72-84. Giersch, C. (2000). Mathematical modelling of metabolism. Curr. Op. Plant Biol. 3, 249-253. Ouzounis, C. A. & Karp, P. D. (2000). Global properties of the metabolic map of Escherichia coli. Genome Research 10, 568-576. Teusink, B., Passarge, J., Reijenga, C. A., Esgalhado, E., van der Weijden, C. C., Schepper, M., Walsh, M. C., Bakker, B. M., van Dam, K., Westerhoff, H. V. & Snoep, J. L. (2000). Can yeast glycolysis be understood in terms of in vitro kinetics of the constituent enzymes? Testing biochemistry. Eur J Biochem 267, 5313-29. Gombert, A. K. & Nielsen, J. (2000). Mathematical modelling of metabolism. Curr. Op. Biotechnol. 11, 180-186. Hofmeyr, J. H. & Westerhoff, H. V. (2001). Building the cellular puzzle: control in multi-level reaction networks. J Theor Biol 208, 261-85. Noble, D. (2002a). The rise of computational biology. Nat. Rev. Mol. Cell Biol. 3, 460-463. Noble, D. (2002b). Modeling the heart – from genes to cells to the whole organ. Science 295, 1678-1682. Hoffmann, A., Levchenko, A., Scott, M. L. & Baltimore, D. (2002). The IkB-NF-kB signaling module: temporal control and selective gene activation. Science 298, 1241-5. Mendes, P. (2002). Emerging bioinformatics for the metabolome. Brief Bioinform 3, 134-45. Kell, D. B. & Westerhoff, H. V. (1986). Metabolic control theory: its role in microbiology and biotechnology. FEMS Microbiol. Rev. 39, 305-320. Bailey, J. E. (1991). Toward a science of metabolic engineering. Science 252, 1668-1675. Hughes, T. R., Marton, M. J., Jones, A. R., Roberts, C. J., Stoughton, R., Armour, C. D., Bennett, H. A., Coffey, E., Dai, H. Y., He, Y. D. D., Kidd, M. J., King, A. M., Meyer, M. R., Slade, D., Lum, P. Y., Stepaniants, S. B., Shoemaker, D. D., Gachotte, D., Chakraburtty, K., Simon, J., Bard, M. & Friend, S. H. (2000). Functional discovery via a compendium of expression profiles. Cell 102, 109-126. Barabasi, A.-L. (2002). Linked: the new science of networks. Perseus Publishing, Cambridge, MA. Watts, D. J. & Strogatz, S. H. (1998). Collective dynamics of 'small-world' networks. Nature 393, 440-2. Wagner, A. & Fell, D. A. (2001). The small world inside large metabolic networks. Proc R Soc Lond B Biol Sci 268, 1803-10. Shen-Orr, S. S., Milo, R., Mangan, S. & Alon, U. (2002). Network motifs in the transcriptional regulation network of Escherichia coli. Nat Genet 31, 64-8. Milo, R., Shen-Orr, S., Itzkovitz, S., Kashtan, N., Chklovskii, D. & Alon, U. (2002). Network motifs: simple building blocks of complex networks. Science 298, 824-7. Ravasz, E., Somera, A. L., Mongru, D. A., Oltvai, Z. N. & Barabasi, A. L. (2002). Hierarchical organization of modularity in metabolic networks. Science 297, 1551-5. von Dassow, G., Meir, E., Munro, E. M. & Odell, G. M. (2000). The segment polarity network is a robust development module. Nature 406, 188-192. Kell, D.B. & Knowles, J.D. (2006) The role of modeling in systems biology. In System modeling in cellular biology: from concepts to nuts and bolts (ed. Z. Szallasi, J. Stelling and V. Periwal), pp. 3-18. MIT Press, Cambridge. Kell, D.B. (2006) Metabolomics, modelling and machine learning in systems biology towards an understanding of the languages of cells. The 2005 Theodor Bücher lecture. The FEBS Journal 273, 873-894. Kell, D.B. (2006) Systems biology, metabolic modelling and metabolomics in drug discovery and development Drug Discovery Today, 11, 1085-1092. |
[lastupdated]





