Welcome
Welcome to our genetic programming and evolutionary computing home page. Use this page to find out more about the genetic programming method and how it is being developed and applied by our group and collaborators.
The Bioanalytical Sciences Group has a very active research on the application of genetic programming and related computational methods to a variety of biological problems, based on the analysis of chemometric data from a variety of analytical instruments. These instruments typically generate data with very high dimensionality, frequently measuring many tens or even hundreds of variables per sample. The goal of much of our work is to derive predictive models by using supervised training of GAs and GPs, with the emphasis on variable selection and meaningful deconvolution (interpretation). Much of our GA/GP research has been done using software developed in-house, which is constantly being refined as new experimental techniques are evolved.
Introduction
Genetic programming [GP] is an optimization technique based on the concepts of Darwinian evolution. A population of individuals, each representing a potential solution to the problem to be optimized, undergoes a process analogous to biological evolution in order to derive an optimal or at least near-optimal solution. The solution offered by each individual is assigned a fitness: a single numerical value which indicates how well that solution performs. New individuals are generated by procedures analogous to biological reproduction, with parents chosen from the existing population with a probability proportional to their fitness. The new individuals may replace less fit members of the population, so the overall population fitness improves with each generation.
Individuals store their potential solutions as a collection of genes. In a genetic algorithm [GA], these may be arrays of bits, integers, floating point numbers, etc., representing parameters of the problem to be optimized, but in a GP they represent program functions, mathematical operators, variables or numerical constants. An individual's total collection of genes is called its genome.
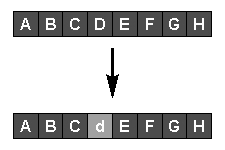
Asexual reproduction, or mutation (see diagram above), is performed by randomly selecting a parent with a probability related to its fitness, then randomly changing one or more genes representing part of the solution it encodes. The fitness of the new individual is assessed, and it replaces a less-fit member of the population if one exists.
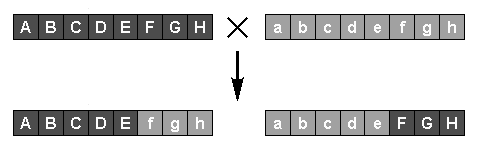
Sexual reproduction, or crossover, is achieved by randomly selecting two parents, again at a rate related to their fitnesses, and generating new individuals by copying genes from one parent, switching to the other at a randomly-selected point. The two new individuals then replace less-fit members of the population as before. The creation of new individuals by mutation and crossover is repeated until an acceptably-fit individual is produced. In a steady state GA only one individual is created per generation; in a generational GA, many new individuals are created at each new generation.
A GP is an application of the GA approach designed to perform an automatic derivation of equations, logical rules or program functions. Rather than representing the solution to the problem as a string of parameters as in a conventional GA, the GP uses a tree structure. The leaves of the tree, or terminals, represent input variables or numerical constants. Their values are passed to nodes, at the junctions of branches in the tree, which perform some arithmetical or program function before passing on the result further towards the root of the tree. Mutations are performed by selecting a parent and either modifying the value or variable returned by a terminal or changing the operation performed by a node. Crossovers are performed by selecting two parents and grafting sub-trees at randomly-selected nodes within their trees. The new individuals so generated again replace less-fit members of the population.
The first implementation of GP, in LISP, was described by John Koza in 1992. More recently, the GP method has been implemented in C/C++, making it more portable between computer platforms. A comprehensive archive of GP source code can be found at the CMU Artificial Intelligence Repository.
| Publications |
|---|
|
Genetic Algorithm Publications: Corne, D. W., Oates, M. J. & Kell, D. B. (2003). Fitness gains and mutation patterns: deriving mutation rates by exploiting landscape data. In Foundations of Genetic Algorithms 7 (ed. K. A. De Jong, R. Poli and J. E. Rowe), pp. 347-364. Morgan Kaufmann, Amsterdam. Corne, D. W., Oates, M. J. & Kell, D. B. (2003). Landscape State Machines: tools for evolutionary algorithm performance analyses and landscape/algorithm mapping. In Evoworkshops 2003, vol. LNCS 2611 (ed. S. Cagnoni, J. R. J. Cardalda, D. W. Corne, J. Gottlieb, A. Guillot, E. Hart, C. G. Johnson, E. Marchiori, J.-M. Meyer, M. Middendorf and G. Raidl), pp. 187-198. Springer, Berlin. Oates, M. J., Corne, D. W. & Kell, D. B. (2003). The bimodal feature at large population sizes and high selection pressure: implications for directed evolution. In Recent advances in simulated evolution and learning (ed. K. C. Tan, M. H. Lim, X. Yao and L. Wang). World Scientific, Singapore. Corne, D.W., Oates, M.J. & Kell, D.B. (2002) On fitness distributions and expected fitness gain of mutation rates in parallel evolutionary algorithms. In Parallel Problem Solving from Nature – PPSN VII. (eds. J.J. Merelo Guervós, P. Adamidis, H.-G. Beyer, J.-L. Fernández-Villacañas & H.-P. Schwefel) 132-141 (Springer, Berlin; 2002). Davies, Z., Gilbert, R.J., Merry, R.J., Kell, D.B., Theodorou, M.K. & Griffith, G.W. (2000) Efficient improvement of silage additives using genetic algorithms. Applied and Environmental Microbiology. 66, 1435-1443. Taylor J., Rowland, J.J., Gilbert, R.J., Jones, A., Winson, M.K. and Kell, D.B. (1998), Genetic Algorithm Decoding for the Interpretation of Infra-red Spectra in Analytical Biotechnology, in Technical Report CSRP-98-10 (EuroGP '98 Late-Breaking Papers), School of Computer Science, University of Birmingham. D. Broadhurst; R. Goodacre; A. Jones; J.J. Rowland and D.B. Kell, Genetic Algorithms as a Method for Variable Selection in PLS Regression with Applications to Pyrolysis Mass Spectrometry, Analytica Chimica Acta 348:71-86 (1997). Genetic Progamming Publications: Vaidyanathan, S., D. I. Broadhurst, D. B. Kell, and R. Goodacre. 2003. Explanatory optimisation of protein mass spectrometry via genetic search. Anal. Chem. 75:6679-6686. Allen, J. K., Davey, H. M., Broadhurst, D., Heald, J. K., Rowland, J. J., Oliver, S. G. & Kell, D. B. (2003). Metabolic footprinting: a high-throughput, high-information approach to cellular characterisation and functional genomics. Nature Biotechnol. 21, 692-696. Kell, D. B. (2002). Genotype-phenotype mapping: genes as computer programs. Trends Genet 18, 555-9. Abstract. Kell, D.B. (2002) Metabolomics and machine learning: explanatory analysis of complex metabolome data using genetic programming to produce simple, robust rules. Mol. Biol. Rep. 29, 237-242. Kell, D. B. (2002). Defence against the flood: a solution to the data mining and predictive modelling challenges of today. Bioinformatics World (part of Scientific Computing News) Issue 1, 16-18. Manuscript version as pdf. Ellis, D. I., Broadhurst, D., Kell, D.B, Rowland, J.J. & Goodacre, R. (2002). Rapid and quantitative detection of the microbial spoilage of meat using Fourier-Transform infrared spectroscopy and machine learning. Appl. Env. Microbiol. 68, 2822-2888. Abstract Pubmed. Abstract ASM. Full text at ASM. Day, J.P., Kell, D.B. & Griffith, G.W. (2002) Differentiation of Phytophthora infestans sporangia from other airborne biological particles by flow cytometry. Appl. Env. Microbiol. 68, 37-45. Abstract. Full version in pdf at AEM. Kell, D.B., Darby, R.M. & Draper, J. (2001) Genomic computing: explanatory analysis of plant expression profiling data using machine learning. Plant Physiology 126, 943.951. Abstract. Full paper at Plant Physiology. Gilbert, R. J., Rowland, J. J. & Kell, D. B. (2000). Genomic computing: explanatory modelling for functional genomics. In Proceedings of the Genetic and Evolutionary Computation Conference (GECCO-2000) (ed. D. Whitley, D. Goldberg, E. Cantú-Paz, L. Spector, I. Parmee and H.-G. Beyer), pp. 551-557. Morgan Kaufmann, San Francisco. Johnson, H.E., Gilbert, R.J., Winson, M.K., Goodacre, R., Smith, A.R., Rowland, J.J., Hall, M.A. & Kell, D.B. (2000). Explanatory analysis of the metabolome using genetic programming of simple, interpretable rules. Genetic Programming and Evolvable Machines 1(3). 243-258. Goodacre, R., Shann, B., Gilbert, R.J., Timmins, É.M., McGovern, A.C., Alsberg, B.K., Kell, D.B. & Logan, N.A. (2000) The detection of the dipicolinic acid biomarker in Bacillus spores using Curie-point pyrolysis mass spectrometry and Fourier-transform infrared spectroscopy. Analytical Chemistry 72, 119-127. Goodacre, R. & Gilbert, R.J. (1999) The detection of caffeine in a variety of beverages using Curie-point pyrolysis mass spectrometry and genetic programming. The Analyst 124, 1069-1074. Woodward, A.M., Gilbert, R.J. and Kell, D.B. (1999). Genetic programming as an analytical tool for non-linear dielectric spectroscopy. Biochemistry and Bioenergetics 48, 389-396. Gilbert, R.J., Johnson, H.E., Winson, M.K., Rowland, J.J., Goodacre, R., Smith, A.R., Hall, M.A. & Kell, D.B. (1999) Genetic programming as an analytical tool for metabolome data. In EuroGP '99 Late-Breaking Papers: Technical Report SEN-R9913 (Eds. Langdon, W.B., Poli, R., Nodin, P. & Fogarty, T.), pp. 23-33. Software Engineering, CWI, Amsterdam. R. Goodacre; B. Shann; R.J. Gilbert; É.M. Timmins; A.C. McGovern; B.K. Alsberg; N.A. Logan and D.B. Kell, The characterisation of Bacillus species from PyMS and FT IR data. In Proceedings of the 1997 ERDEC Scientific Conference on Chemical and Biological Defense Research. ERDEC-SP-063, Aberdeen Proving Ground (1999). Jones, A., Young, D., Taylor, J., Kell, D.B. & Rowland, J.J. (1998) Quantification of microbial productivity via multi-angle light scattering and supervised learning. Biotechnol. Bioeng., 59, 131-143. Abstract. R.J. Gilbert; R. Goodacre; B. Shann; J. Taylor; J.J. Rowland and D.B. Kell, Genetic Programming-Based Variable Selection for High-Dimensional Data, Genetic Programming 1998: Proceedings of the Third Annual Conference (J.R. Koza et al., eds.), Morgan Kaufmann (1998). J. Taylor; M.K. Winson; R. Goodacre; R.J. Gilbert; J.J. Rowland and D.B. Kell, Genetic Programming in the Interpretation of Fourier-Transform Infrared Spectra: Quantification of Metabolites of Pharmaceutical Importance, Genetic Programming 1998: Proceedings of the Third Annual Conference (J.R. Koza et al., eds.), Morgan Kaufmann, San Fransisco CA (1998). Taylor, J., Goodacre, R., Wade, W.G., Rowland, J.J. & Kell, D.B. (1998) The deconvolution of pyrolysis mass spectra using genetic programming: application to the identification of some Eubacterium species. FEMS Microbiology Letters 160, 237- 246. Abstract. Gilbert, R.J., Goodacre, R., Woodward, A.M. & Kell, D.B. (1997) Genetic programming: a novel method for the quantitative analysis of pyrolysis mass spectral data. Anal. Chem. 69, 4381-4389. |
| Links |
|---|
|
Some useful Genetic Programming links… Very much under construction… People Bibliographic:
The GP bibliography Tutorials:
Genetic Programming FAQs
Another one in the same place Hardware:
Adrian Stoica JPL Software:
Academic Other overviews: Parallel and Evolutionary Computing on Beowulf Class supercomputers: |
[lastupdated]





